Chapter 11 Assignment 1: Basic Programming in R
Report the following results in html format using R markdown. In other words, replicate this document. You write functions in a separate R file and put in R folder in the project folder. Build the project as a package and load it from the R markdown file. The execution code sholuld be written in R markdown file.
You submit:
- R file containing functions.
- R markdown file containing your answers and executing codes.
- HTML(or PDF) report generated from the R markdown.
11.1 Simulate data
Consider to simulate data from the following model and estimate the parameters from the simulated data.
\[ y_{ij} = 1\{j = \text{argmax}_{k = 1, 2} \beta x_k + \epsilon_{ik} \}, \] where \(\epsilon_{ik}\) follows i.i.d. type-I extreme value distribution, \(\beta = 0.2\), \(x_1 = 0\) and \(x_2 = 1\).
- To simulate data, first make a data frame as follows:
## # A tibble: 2,000 × 3
## i k x
## <int> <int> <dbl>
## 1 1 1 0
## 2 1 2 1
## 3 2 1 0
## 4 2 2 1
## 5 3 1 0
## 6 3 2 1
## 7 4 1 0
## 8 4 2 1
## 9 5 1 0
## 10 5 2 1
## # ℹ 1,990 more rows- Second, draw type-I extreme value random variables. Set the seed at 1. You can use
evdpackage to draw the variables. You should get exactly the same realization if the seed is correctly set.
## # A tibble: 2,000 × 4
## i k x e
## <int> <int> <dbl> <dbl>
## 1 1 1 0 0.281
## 2 1 2 1 -0.167
## 3 2 1 0 1.93
## 4 2 2 1 1.97
## 5 3 1 0 0.830
## 6 3 2 1 -1.06
## 7 4 1 0 -0.207
## 8 4 2 1 0.617
## 9 5 1 0 0.0444
## 10 5 2 1 1.92
## # ℹ 1,990 more rows- Third, compute the latent value of each option to obtain the following data frame:
## # A tibble: 2,000 × 5
## i k x e latent
## <int> <int> <dbl> <dbl> <dbl>
## 1 1 1 0 0.281 0.281
## 2 1 2 1 -0.167 0.0331
## 3 2 1 0 1.93 1.93
## 4 2 2 1 1.97 2.17
## 5 3 1 0 0.830 0.830
## 6 3 2 1 -1.06 -0.863
## 7 4 1 0 -0.207 -0.207
## 8 4 2 1 0.617 0.817
## 9 5 1 0 0.0444 0.0444
## 10 5 2 1 1.92 2.12
## # ℹ 1,990 more rows- Finally, compute \(y\) by comparing the latent values of \(k = 1, 2\) for each \(i\) to obtain the following result:
## # A tibble: 2,000 × 6
## i k x e latent y
## <int> <int> <dbl> <dbl> <dbl> <dbl>
## 1 1 1 0 0.281 0.281 1
## 2 1 2 1 -0.167 0.0331 0
## 3 2 1 0 1.93 1.93 0
## 4 2 2 1 1.97 2.17 1
## 5 3 1 0 0.830 0.830 1
## 6 3 2 1 -1.06 -0.863 0
## 7 4 1 0 -0.207 -0.207 0
## 8 4 2 1 0.617 0.817 1
## 9 5 1 0 0.0444 0.0444 0
## 10 5 2 1 1.92 2.12 1
## # ℹ 1,990 more rows11.2 Estimate the parameter
Now you generated simulated data. Suppose you observe \(x_k\) and \(y_{ik}\) for each \(i\) and \(k\) and estimate \(\beta\) by a maximum likelihood estimator. The likelihood for \(i\) to choose \(k\) (\(y_{ik} = 1\)) can be shown to be: \[ p_{ik}(\beta) = \frac{\exp(\beta x_k)}{\exp(\beta x_1) + \exp(\beta x_2)}. \]
Then, the likelihood of observing \(\{y_{ik}\}_{i, k}\) is: \[ L(\beta) = \prod_{i = 1}^{1000} p_{i1}(\beta)^{y_{i1}} [1 - p_{i1}(\beta)]^{1 - y_{i1}}, \] and the log likelihood is: \[ l(\beta) = \sum_{i = 1}^{1000}\{y_{i1}\log p_{i1}(\beta) + (1 - y_{i1})\log [1 - p_{i1}(\beta)\}. \]
- Write a function to compute the likelihood for a given \(\beta\) and data and name the function
compute_loglikelihood_a1(b, df).
## [1] -0.7542617- Compute the value of log likelihood for \(\beta = 0, 0.1, \cdots, 1\) and plot the result using
ggplot2packages. You can uselatex2exppackage to use LaTeX math symbol in the label:
b_seq <- seq(0, 1, 0.1)
output <-
foreach (
b = b_seq,
.combine = "rbind"
) %do% {
l <-
compute_loglikelihood_a1(
b,
df
)
return(l)
}
output <-
data.frame(
x = b_seq,
y = output
)
output %>%
ggplot(
aes(
x = x,
y = y
)
) +
geom_point() +
xlab(TeX("$\\beta$")) +
ylab("Loglikelihood") +
theme_classic()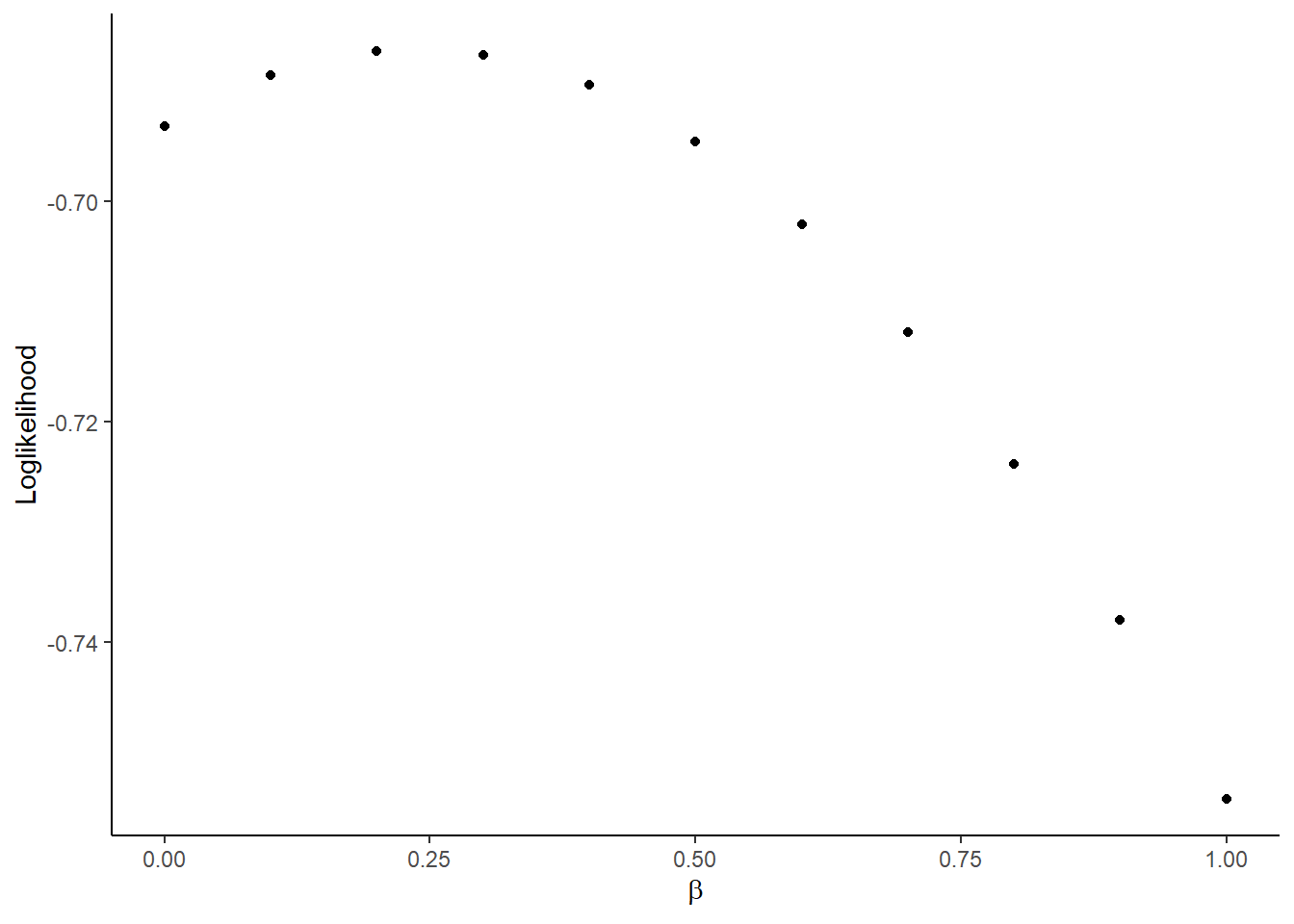
- Find and report \(\beta\) that maximizes the log likelihood for the simulated data. You can use
optimfunction to achieve this. You will useBrentmethod and set the lower bound at -1 and upper bound at 1 for the parameter search.
## $par
## [1] 0.2371046
##
## $value
## [1] -0.6861689
##
## $counts
## function gradient
## NA NA
##
## $convergence
## [1] 0
##
## $message
## NULL